CHIP develops, advances, and applies computational tools to analyze genomic data, essential for understanding and addressing severe chronic diseases such as cancer, neurodegenerative conditions, and rare genetic disorders. Utilizing next-generation sequencing, CHIP examines genomic aberrations in both affected cells and germline DNA. Through initiatives like the Genomics Information Commons (GIC), CHIP connects pediatric hospitals across the U.S. to identify disease-causing genetic variants and support multi-omics approaches, including transcriptomics, proteomics, and phenomics from electronic health records. By integrating these data with deep learning techniques, CHIP enhances understanding of disease mechanisms and fosters innovative therapies, including personalized “N-of-1” clinical trials
Projects

Inspired by a common vision of accelerated genomic discovery, collaboration, and improved clinical outcomes, leaders at Boston Children's Hospital, Cincinnati Children's Hospital Medical Center, the Children's Hospital of Philadelphia, Washington University at St. Louis, the University of Pittsburgh Medical Center, and Le Bonheur Children's Hospital at the University of Tennessee Health Science Center have come together to create the Genomic Information Commons (GIC). Partially funded by NCATS, The GIC is a multi-institutional, federated, genomic data commons.

CHIP directs the Biobank Core at Boston Children’s Hospital which includes the Precision Link Health Discovery protocol for engaging our patients in research and capturing the diversity of our patient population under a broad consent for omics and phenotype data phenotype biospecimens stored in the biobank core.
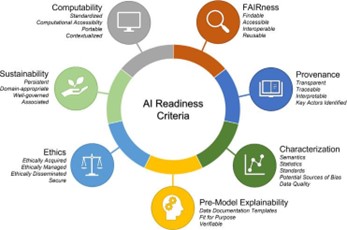
Our team at CHIP is working to establish robust data standards that will enable AI/ML methods to analyze and predict genotype-phenotype associations more effectively. As a part of the groundbreaking NIH-funded program called “Bridge to Artificial Intelligence (Bridge2AI), we are advancing biomedical data to support AI/ML analysis. A key focus of Bridge2AI is the development of standards for genomic variants, metadata, and phenotype information. Our work with Bridge2AI will not only enhance the accuracy of research, but also lays the foundation for improved diagnostics, treatment, and personalized medicine.
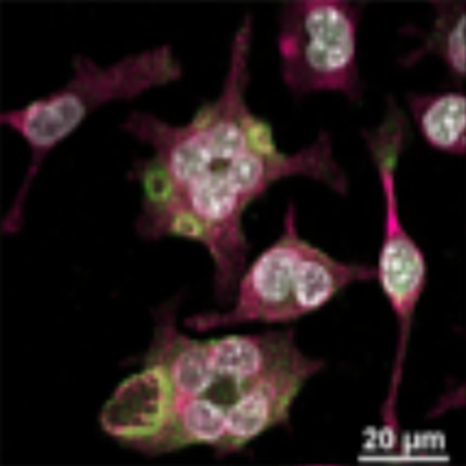
Microglia, the brain’s resident immune cells, play a crucial role in clearing harmful proteins and debris that accumulate with age and disease and can contribute to the buildup of amyloid-beta plaques and other key features of Alzheimer’s disease. Recent research has focused on the Pyk2 enzyme, which genetic studies have linked to Alzheimer’s. Targeting Pyk2 may enhance microglial function, potentially reducing amyloid-beta plaque formation and slowing disease progression. Our team at CHIP uses both in vitro and in vivo model systems to test small molecules that inhibit Pyk2 activity. Initial findings suggest that carefully modulating microglial activity through Pyk2 inhibition may help preserve brain health and prevent or delay Alzheimer’s disease and related dementias.
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, primarily caused by the loss of functional dopaminergic neurons in the brain. A promising therapeutic approach involves transplanting dopaminergic neurons derived from a patient’s own cells. We have successfully treated a patient with PD using this strategy. However, a critical challenge remains: generating clinical-grade induced pluripotent stem cell lines free of potentially harmful somatic mutations. Our team is focused on establishing and refining protocols that maintain genome integrity and ensure mutation-free cell lines for future clinical trials. This work aims to advance the safety and effectiveness of autologous cell replacement therapies for PD.
The Clinical Pharmacogenomics Service (PGx Clinic) works to make medications safer and more effective by incorporating patient’s genetic information when making decisions about medication choices. The PGx Clinic applies genetics to determine variations in medication response to improve clinical care.

Autism is caused by gene x environment interaction during brain development. We aim to investigate what contributed a significant increase to the autism prevalence in the U.S. over the past decades, focusing on discovery of internal and external environmental exposures that are detectable in the blood from the patients with autism and their family members compared to individuals without autism.
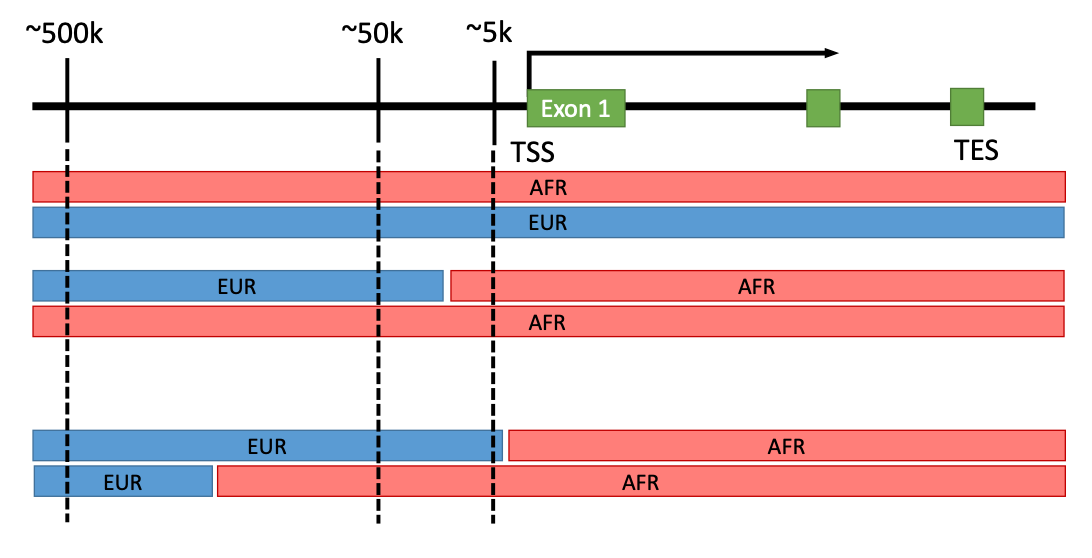
Regulation of gene expression is complex and non-coding regions of human genome have diverse regulatory elements. We hypothesize that local ancestry specific haplotypes containing regulatory elements have impacts on (local-) ancestry specific difference in gene expression regulation. 70% of human genome may include regulatory elements of which some are unique to ancestry-specific haplotypes.
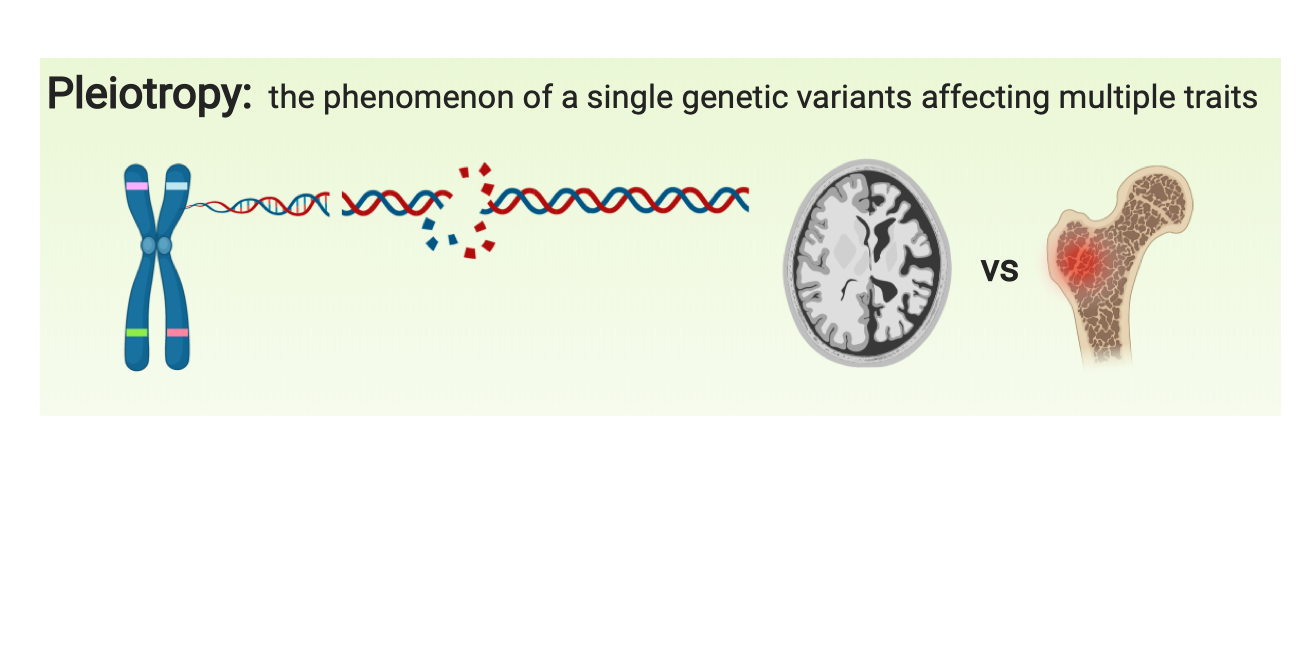
Tissue-resident macrophages are highly specialized to their tissue-specific microenvironments and activated by various inflammatory signals and modulated by genetic and environmental factors. Osteoclasts and microglia are distinct tissue-resident macrophages for bone and brain that are responsible for pathological changes in osteoporosis and Alzheimer’s disease (AD), respectively. Pleiotropy is the phenomenon that a genetic variant affects multiple traits. We discover the genes with pleiotropy affecting brain and bone by focusing on tissue-resident macrophages.

Synthetic lethality based therapeutic strategies targeting specific DNA repair deficiencies have become an effective treatment modality in solid tumors. For example, PARP inhibitors provide significant clinical benefit in homologous recombination deficient, such as BRCA1 or BRCA2 mutant, breast, ovarian or prostate cancer. The success of such therapies, however, depends on the diagnostic detection of the particular DNA repair deficiency present in the cancer, which can be achieved by the analysis of specific mutational profiles of the tumor DNA. CHIP researchers have developed clinically useful diagnostic mutational signatures of homologous recombination deficiency and nucleotide excision repair deficiency, which are used to prioritize patients for PARP inhibitors and other novel therapies such as derivatives of irofulven.




